Swiss Institute of Bioinformatics
Click2Drug
Directory of computer-aided Drug Design tools
Click2Drug contains a comprehensive list of computer-aided drug design (CADD) software, databases and web services. These tools are classified according to their application field, trying to cover the whole drug design pipeline. If you think that an interesting tool is missing in this list, please contact usClick on the following picture to select tools related to a given activity:
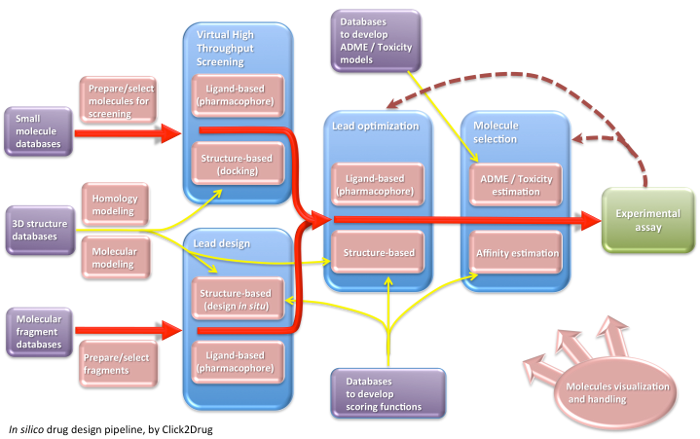
Show all links Hide all links
ADME Toxicity
Databases
- PACT-F. (Preclinical And Clinical Trials Knowledge Base on Bioavailability). Preclinical And Clinical Trials Knowledge Base on Bioavailability (PACT-F). The database contains 8296 records, which describe in detail the results of clinical trials in humans and preclinical trials in animals. PACT-F is extensively annotated. Up to 17 fields describe in detail the results and conditions of each trial, such as route of administration, species investigated, drug formulation, coadministration of drug, feeding condition, age and gender of the subjects involved, dosing scheme, genetic differences, experimental and analytical procedure, method of calculation and state of health. Provided by PharmaInformatic, Germany.
- TOXNET. Databases on toxicology, hazardous chemicals, environmental health, and toxic releases that can be accessed using a common search interface. provided by the Unied States NLM.
- Leadscope Toxicity Database. Database of 160,000 chemical structures with toxicity data. Distributed by Leadscope.
- WOMBAT-PK. Database for Clinical Pharmacokinetics and Drug Target Information. WOMBAT-PK contains 1260 entries (1260 unique SMILES), totaling over 9,450 clinical pharmacokinetic measurements; it further includes 2,316 physico-chemical properties; 932 toxicity endpoints, and 2,186 annotated drug-target bioactivities. Compiled by Sunset Molecular Discovery LLC.
- Cloe Knowledge. Open Access ADME/PK Database for a range of marketed drugs. Maintained by Cyprotex.
- PHYSPROP. The Physical Properties Database (PHYSPROP) contains chemical structures, names, and physical properties for over 41,000 chemicals. Physical properties are collected from a wide variety of sources, and include experimental, extrapolated, and estimated values for melting point, boiling point, water solubility, octanol-water partition coefficient, vapor pressure, pKa, Henry's Law Constant, and OH rate constant in the atmosphere. Maintained by SRC.
- SIDER. Contains information on marketed medicines and their recorded adverse drug reactions. The information is extracted from public documents and package inserts. The available information include side effect frequency, drug and side effect classifications as well as links to further information, for example drug–target relations. Provided by the European Molecular Biology Laboratory, Heidelberg, Germany.
- admetSAR. admetSAR provides the manually curated data for diverse chemicals associated with known Absorption, Distribution, Metabolism, Excretion and Toxicity profiles. admetSAR allows searching for ADMET properties profiling by name, CASRN and similarity search. In addition, admetSAR can predict about 50 ADMET endpoints by our recently development chemoinformatics-based toolbox, entitled ADMET-Simulator.
- The ADME databases. Databases for benchmarking the results of experiments, validating the accuracy of existing ADME predictive models, and building new predictive models.
- The ADME database. Provides comprehensive data for structurally diverse compounds associated with known ADME properties, including human oral bioavailability, enzymes metabolism, inhibition and induction, transport, plasma protein binding and bloodbrain barrier. Distributed by Aureus.
- UCSF-FDA Transportal. The purpose of this database is to be a useful repository of information on transporters important in the drug discovery process as a part of the US Food and Drug Administration-led Critical Path Initiative. Information includes transporter expression, localization, substrates, inhibitors, and drug-drug interactions It contains 3438 compounds, 11649 interaction records, 1211 literature references. The FDA has partnered with the Giacomini lab at UCSF to create a transporter database of pharmacologically relevant transporters to support development of new pharmaceuticals. Information on important transporters, their localization, expression levels, substrates, and inhibitors have been curated from the literature and compiled into a single location to aid and inform drug developers, regulatory agencies and academic scientists about transporters important in drug action and disposition.. The database will also help drug developers in determining what experiments or analyses must be conducted to check for possible drug interactions through transporters as well as identify promising transporter candidates for the testing of possible genetic influences.
- SuperTarget Database. Database of about 332828 drug-target relations.
- DART. (Drug Adverse Reaction Target). A database for facilitating the search for drug adverse reaction target. It contains information about known drug adverse rection targets, functions and properties. Associated references are also included. Maintained by the University of Singapore.
- DITOP. (Drug-Induced Toxicity Related Proteins). Database of proteins that mediate toxicities through their interaction with drugs or reactive metabolites. Can be searched using keywords of chemicals, proteins, or toxicity terms. Maintained by the Xiamen university.
- ADMEAP. A database for facilitating the search for drug Absorption, Distribution, Metabolism, Excretion associated proteins. It contains information about known drug ADME associated proteins, functions, similarities, substrates / ligands, tissue distributions, and other properties of the targets. Associated references are also included. Currently this database contains 321 protein entries. Maintained by the Dept.Computational Science. NUS.
- SIDER. (Side Effect Resource). contains information on marketed medicines and their recorded adverse drug reactions. The information is extracted from public documents and package inserts. The available information include side effect frequency, drug and side effect classifications as well as links to further information, for example drug–target relations.
- SAR Genetox Database. Genetic toxicity database to be used as a resource for developing predictive modeling training sets. Distributed by Leadscope.
- SAR Carcinogenicity Database. Carcinogenicity database with validated structures to be used as a resource for preparing training sets. Distributed by Leadscope.
- HMDB. The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. The database contains chemical data, clinical data, and molecular biology/biochemistry data. The database (version 2.5) contains over 7900 metabolite entries including both water-soluble and lipid soluble metabolites as well as metabolites that would be regarded as either abundant (> 1 uM) or relatively rare (< 1 nM). Provided by the Departments of Computing Science & Biological Sciences, University of Alberta.
- t3db. (Toxin and Toxin Target Database). Combines detailed toxin data with comprehensive toxin target information. The database currently houses over 2900 toxins described by over 34 200 synonyms, including pollutants, pesticides, drugs, and food toxins, which are linked to over 1300 corresponding toxin target records. Altogether there are over 33 800 toxin, toxin target associations. Each toxin record (ToxCard) contains over 50 data fields and holds information such as chemical properties and descriptors, toxicity values, molecular and cellular interactions, and medical information. This information has been extracted from over 5600 sources, which include other databases, government documents, books, and scientific literature. Provided by the Departments of Computing Science & Biological Sciences, University of Alberta.
- SuperToxic. Collection of toxic compounds from literature and web sources. The current version of this database compiles approx. 60,000 compounds with about 100,000 synonyms. These molecules are classified according to their toxicity based on more than 2,500,000 measurements. Provided by Charité Berlin, Structural Bioinformatics Group.
- SuperHapten. Comprehensive database for small immunogenic compounds. Contains currently 7257 haptens, 453 commercially available related antibodies and 24 carriers. Provided by Charité Berlin, Institute of Molecular Biology and Bioinformatics.
- HaptenDB. Database of about 1087 haptens that includes common and chemical name of Hapten, molecular mass, physical and chemical properties, biological importance and the structure. Provided by the Institute of Microbial Technology, India.
- SuperCyp. Comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Provided by Charité Berlin, Structural Bioinformatics Group.
- PROMISCUOUS. Exhaustive resource of protein-protein and drug-protein interactions with the aim of providing a uniform data set for drug repositioning and further analysis. PROMISCUOUS contains three different types of entities: drugs, proteins and side-effects as well as relations between them. Provided by Charité Berlin, Structural Bioinformatics Group.