Swiss Institute of Bioinformatics
Click2Drug
Directory of computer-aided Drug Design tools
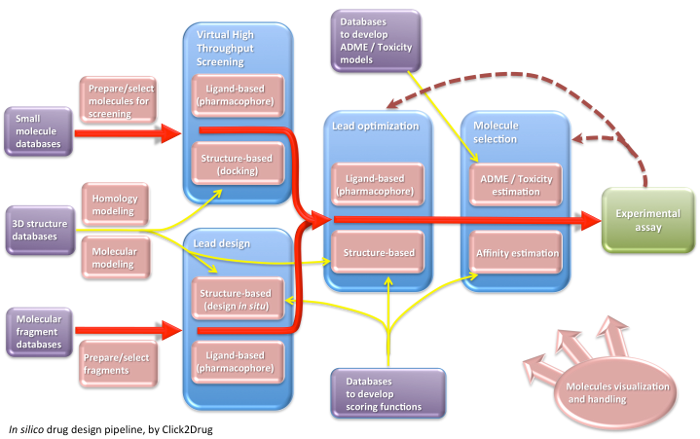
Click2Drug contains a comprehensive list of computer-aided drug design (CADD) software, databases and web services. These tools are classified according to their application field, trying to cover the whole drug design pipeline. If you think that an interesting tool is missing in this list, please contact usClick on the following picture to select tools related to a given activity:
Show all links Hide all links
Binding site prediction
Software
- MED-SuMo. Program for macromolecules surface similarity detection. Searches into 3D databases, find similar binding surfaces and generate 3D superpositions based on common surface chemical features and similar shape. Can be used for site mining, drug repurposing and site classification at PDB scale. Distributed by MEDIT.
- TRAPP. TRAnsient Pockets in Proteins (TRAPP) is a web server for the analysis of transient binding pockets in proteins. Contrarily to many tools, it is not intended for ligand binding pocket identification per se, but rather to predict significant changes in the spatial and physicochemical properties of a given pocket that may arise due to the protein's flexibility (both backbone and side chains). Several capabilities of visualization and analysis have been developed and are provided by the Molecular and Cellular Modeling group at Heidelberg Institute for Theoretical Studies, Germany.
- CAVER. Software tool for analysis and visualisation of tunnels and channels in protein structures. Provided by the Masaryk University.
- fpocket. Open source protein pocket (cavity) detection algorithm based on Voronoi tessellation. Developed in the C programming language and currently available as command line driven program. fpocket includes two other programs (dpocket & tpocket) that allow you to extract pocket descriptors and test own scoring functions respectively. Also contains a druggability prediction score.
- GHECOM. Program for finding multi-scale pockets on protein surfaces using mathematical morphology. Free open source.
- LIGSITEcsc. Program for the automatic identification of pockets on protein surface using the Connolly surface and the degree of conservation.
- SURFNET. Generates surfaces and void regions between surfaces from coordinate data supplied in a PDB file.
- SiteHound. Identifies ligand binding sites by computing interactions between a chemical probe and a protein structure. The input is a PDB file of a protein structure, the output is a list of “interaction energy clusters” corresponding to putative binding sites.
- ICM-PocketFinder. Binding site predictor based on calculating the drug-binding density field and contouring it at a certain level. Provided by Molsoft.
- SiteMap. Program for binding site identification. Distributed by Schrodinger.
- MSPocket. Orientation independent program for the detection and graphical analysis of protein surface pockets. A MSPocket plugin for PyMOL provides a graphical user interface for runing MSPocket and render its results in PyMOL. It is included in the download. Free and open source.
- POCASA. (POcket-CAvity Search Application). Automatic web service that implements the algorithm named Roll which can predict binding sites by detecting pockets and cavities of proteins of known 3D structure. Maintained by the Hokkaido University.
- Phosfinder. Method for the prediction of phosphate-binding sites in protein structures. provided by the University of Rome.
- VOIDOO. Software to find cavities and analyse volumes.
- FunFOLDQA. Program to assess the quality ligand binding site residue predictions based on 3D models of proteins. Free program written in java. Developped by the University of Reading.
- LISE. Free and open source program for ligand Binding Site Prediction Using Ligand Interacting and Binding Site-Enriched Protein Triangles. Exists as a web service. Provided by the Institute of Biomedical Sciences, Academia Sinica.
- PDBinder. Free program for the identification of small ligand-binding sites in a protein structure. webPDBinder searches a protein structure against a library of known binding sites and a collection of control non-binding pockets. Exists as a web service. Provided by the University of Roma 2, Italy.
- eFindSite. Ligand binding site prediction and virtual screening algorithm that detects common ligand binding sites in a set of evolutionarily related proteins identified by 10 threading/fold recognition methods. Exists as a web service. Provided by the Louisiana State University, Computational Systems Biology Group.
- POVME. Free and open source program for measuring binding-pocket volumes. Developed by the National Biomedical Computation Resource.
- SiteEngine. Program to predict regions that can potentially function as binding sites. The methods is based on recognition of geometrical and physico-chemical environments that are similar to known binding sites. Exists as a web service. Provided by the structural Bioinformatics group at Tel-Aviv University.
- SVILP_ligand. General method for discovering the features of binding pockets that confer specificity for particular ligands. Provided by the Computational Bioinformatics Laboratory, Imperial College London.
Databases
- sc-PDB. Annotated Database of Druggable Binding Sites from the Protein DataBank. Provided by the university of Strasbourg.
- CASTp. Computed Atlas of Surface Topography of proteins. Provides identification and measurements of surface accessible pockets as well as interior inaccessible cavities, for proteins and other molecules. castP server uses the weighted Delaunay triangulation and the alpha complex for shape measurements.
- Pocketome. Encyclopedia of conformational ensembles of all druggable binding sites that can be identified experimentally from co-crystal structures in the Protein Data Bank.
- PDBe motifs and Sites. Can be used to examine the characteristics of the binding sites of single proteins or classes of proteins such as Kinases and the conserved structural features of their immediate environments either within the same specie or across different species.
- LigASite. Dataset of biologically relevant binding sites in protein structures. It consists of proteins with one unbound structure and at least one structure of the protein-ligand complex. Both a redundant and a non-redundant (sequence identity lower than 25%) version is available.
- PROtein SURFace ExploreR. Contains information about structural similarities with respect to the query surfaces. A pocket search algorithm detected 48,347 potential ligand binding sites from the 9,708 non-redundant protein entries in the PDB database. All-against-all structural comparison was performed for the predicted sites, and the similar sites with the Z-score ≥ 2.5 were selected. These results can be accessed by the PDB code or ligand name.
- fPOP. Footprinting protein functional surfaces by comparative spatial patterns. Database of the protein functional surfaces identified by shape analysis.
- PDBSITE. Database on protein active sites and their spatial environment. Provided by GeneNetworks.
- LigBase. Database of ligand binding proteins aligned to structural templates. The structural templates are taken from the PDB, 3D models of the aligned sequences are provided ModBase, and pairwise sequence alignments are provided by CE. Multiple Structural Alignments are built on the fly within LigBase from a series of pairwise alignments. Ligand diagrams are generated with the program Ligplot. Maintained by Andrej Sali at the University of California, San Francisco.
Web services
- 3DLigandSite. Automated method for the prediction of ligand binding sites. Provided by the Imperial London College.
- metaPocket. Meta server to identify pockets on protein surface to predict ligand-binding sites.
- PockDrug. A methodology tehat predicts pocket druggability, efficient on both; estimated pockets guided by the ligand proximity (extracted by proximity to a ligand from a holo protein structure using several thresholds) and estimated pockets not guided by the ligand proximity (based on amino atoms that form the surface of potential binding cavities).. Developed and maintained by the University Paris-Diderot, France.
- PocketQuery. Protein-protein interaction (PPI) inhibitor starting points from PPI structure. Quickly identify a small set of residues at a protein interface that are suitable starting points for small-molecule design. Provided by the University of Pittsburgh.
- PASS. Program for tentative identification of drug interaction pockets from protein structure.
- DEPTH. Web server to compute depth and predict small-molecule binding cavities in proteins
- fpocket web server. Open source protein pocket (cavity) detection algorithm based on Voronoi tessellation. Developed in the C programming language and currently available as command line driven program. fpocket includes two other programs (dpocket & tpocket) that allow you to extract pocket descriptors and test own scoring functions respectively. Also contains a druggability prediction score.
- Nucleos. Nucleos is a webserver for the identification of nucleotide-binding sites based on the concept of nucleotide modularity. Nucleos identifies binding sites for nucleotide modules (namely the nucleobase, the carbohydrate and the phosphate) and then combines them in order to build the complete binding sites for different types of nucleotides (e.g. ADP or FAD). Provided by the University of Roma 2, Italy.
- wwwPDBinder. Web server for the identification of small ligand-binding sites in a protein structure. webPDBinder searches a protein structure against a library of known binding sites and a collection of control non-binding pockets. Exists as a standalone program. Provided by the University of Roma 2, Italy.
- IsoMIF. IsoMIF identifies binding site molecular interaction field similarities between proteins. The IsoMIF Finder Interface allows you to identify binding site molecular interaction field (MIF) similarities between a query structure and a database of pre-calculated MIFs or you own custom PDB entries. Developed by the University of Sherbrooke, Canada.
- LISE. Ligand Binding Site Prediction Using Ligand Interacting and Binding Site-Enriched Protein Triangles. Exists as a standalone program. Provided by the Institute of Biomedical Sciences, Academia Sinica.
- eFindSite. Web server for ligand binding site prediction and virtual screening algorithm that detects common ligand binding sites in a set of evolutionarily related proteins identified by 10 threading/fold recognition methods. Exists as standalone program. Provided by the Louisiana State University, Computational Systems Biology Group.
- Active Site Prediction. Web server for computing the cavities in a given protein. Provided by the Supercomputing Facility for Bioinformatics & Computational Biology, IIT Delhi.
- GHECOM web server. Web server for finding multi-scale pockets on protein surfaces using mathematical morphology.
- LIGSITEcsc web server. Web server for the automatic identification of pockets on protein surface using the Connolly surface and the degree of conservation.
- ProBis. Web server for detection of structurally similar binding sites. Maintained by the National Institute of Chemistry, Ljubljana, Slovenia.
- ProBiS-CHARMMing. Web server for detection of structurally similar binding sites, plus minimization of predicted protein-ligand complexes and their interaction energy calculation. Maintained by the National Institute of Health, USA.
- FunFOLD. Web server to predict likely ligand binding site residues for a submitted amino acid sequence.
- CAVER. Software tool for analysis and visualisation of tunnels and channels in protein structures. Provided by the Masaryk University.
- SuMo. Screens the Protein Data Bank (PDB) for finding ligand binding sites matching your protein structure or inversely, for finding protein structures matching a given site in your protein. Provided freely by the Pole Bioinformatique Lyonnais.
- IBIS. (Inferred Biomolecular Interactions Server). For a given protein sequence or structure query, IBIS reports physical interactions observed in experimentally-determined structures for this protein. IBIS also infers/predicts interacting partners and binding sites by homology, by inspecting the protein complexes formed by close homologs of a given query.
- PocketDepth. Depth based algortihm for identification of ligand binding sites.
- Screen2. Tool for identifying protein cavities and computing cavity attributes that can be applied for classification and analysis.
- SiteHound-web. Identifies ligand binding sites by computing interactions between a chemical probe and a protein structure. The input is a PDB file of a protein structure, the output is a list of “interaction energy clusters” corresponding to putative binding sites. Maintained by the Sanchez lab, at the Mount Sinai School of Medicine, NY, USA.
- SiteComp. Web server providing three major types of analysis based on molecular interaction fields: binding site comparison, binding site decomposition and multi-probe characterization. Maintained by the Sanchez lab, at the Mount Sinai School of Medicine, NY, USA.
- ConCavity. Ligand binding site prediction from protein sequence and structure.
- SplitPocket. Prediction of binding sites for unbound structures.
- PepSite 2. Web service for the prediction of peptide binding sites on protein surfaces. Developed and maintained by the Russel Lab, University of Heidelberg.
- MolAxis. Web server for the identification of channels in macromolecules.
- PDBSiteScan. Tool for search for functional sites in protein tertiary structures. Developed in collaboration with Institute of Cytology and Genetics, Novosibirsk.
- MultiBind. (Multiple Alignment of Protein Binding Sites). Prediction tool for protein binding sites. Users input a set of protein-small molecule complexes and MultiBind predicts the common physio-chemical patterns responsible for protein binding. Exists as a standalone program. Provided by the structural Bioinformatics group at Tel-Aviv University.
- SiteEngine. Web service to predict regions that can potentially function as binding sites. The methods is based on recognition of geometrical and physico-chemical environments that are similar to known binding sites. Exists as a standalone program. Provided by the structural Bioinformatics group at Tel-Aviv University.
Docking
Software
- Computer-Aided Drug-Design Platform using PyMOL. PyMOL plugins providing a graphical user interface incorporating individual academic packages designed for protein preparation (AMBER package and Reduce), molecular mechanics applications (AMBER package), and docking and scoring (AutoDock Vina and SLIDE).
- Autodock Vina plugin for PyMOL. Allows defining binding sites and export to Autodock and VINA input files, doing receptor and ligand preparation automatically, starting docking runs with Autodock or VINA from within the plugin, viewing grid maps generated by autogrid in PyMOL, handling multiple ligands and set up virtual screenings, and set up docking runs with flexible sidechains.
- GriDock. Virtual screening front-end for AutoDock 4. GriDock was designed to perform the molecular dockings of a large number of ligands stored in a single database (SDF or Zip format) in the lowest possible time. It take the full advantage of all local and remote CPUs through the MPICH2 technology, balancing the computational load between processors/grid nodes. Provided by the Drug Design Laboratory of the University of Milano.
- DockoMatic. GUI application that is intended to ease and automate the creation and management of AutoDock jobs for high throughput screening of ligand/receptor interactions.
- BDT. Graphic front-end application which control the conditions of AutoGrid and AutoDock runs. Maintained by the Universitat Rovira i Virgili,
Web services
- PLATINUM. Calculates hydrophobic properties of molecules and their match or mismatch in receptor–ligand complexes. These properties may help to analyze results of molecular docking.
Screening
Software
- MedChem Studio. Cheminformatics platform for computational and medicinal chemists supporting lead identification and optimization, in silico ligand based design, and clustering/classifying of compound libraries. It is integrated with MedChem Designer and ADMET Predictor. Distributed by Simulation Plus, Inc.
Ligand design
Software
- GANDI. Program for structure-based fragment-based ab initio (de novo) ligand design. Developed and distributed by the Computational Structural Biology group of prof. Amedeo Caflisch, Zurich University.
- LUDI. Program for automated structure-based drug design, using growing and linking approaches. Distributed by Accelrys as part of Discovery Studio.
- AutoT&T2. The Automatic Tailoring and Transplanting (AutoT&T) method is developed as a versatile computational tool for lead optimization as well as lead discovery in molecular-targeted drug design. This method detects suitable fragments on reference molecules, e.g. outputs of a virtual screening job in prior, and then transplants them onto the given lead compound to generate new ligand molecules. Then, binding affinities, synthetic feasibilities and drug-likeness properties are evaluated to select the promising candidates for further consideration. Standalone software and demo web version
- Allegrow. Program for structure-based fragment-based ligand design, based on growing and combinatorial approaches. Distributed by Boston De Novo Design.
- E-novo. Program for automated structure-based ligand design, using a combinatorial substitution of R-groups on the initial scaffold. Distributed by Accelrys as part of Discovery Studio.
- SILCS. The Site Identification by Ligand Competitive Saturation Method (SILCS) allows for location and estimation of affinities of chemical groups on a protein surface. It relies on MD of the macromolecular structure in an explicit solvent/small molecules environment, representative of chemical fragments of diverse properties (hydophobic, aromatic, with H-bonding capacity). Maps are so created indidcating favorable fragment-protein interactions, to be used for structure-based designed. Distributed by SilcsBio, LLC.
- BOMB. Program for structure-based fragment-based ligand design, based on a growing approach. Distributed by Cemcomco.
- ChIP. Program, based on a genetic algorithm, for the exploration of synthetically feasible small molecule chemical space from commercially available starting materials, directly toward medicinally relevancy, applying predictive computational QSAR models and physicochemical and structural filters. ChIP can take account of propriatary chemical reactions. Developed by Eidogen-Sertanty, Inc.
- ChemT. Open-source software for building chemical compound libraries, based on a specific chemical template. The compound libraries generated can then be evaluated, using several Virtual Screening tools like molecular docking or QSAR modelling tools. Distributed by BioChemCore.
- MEGA. Program for structure-based fragment-based ligand design, based on a EA approach. Distributed by Noesis Informatics (NSisToolkit).
- LigBuilder. Program for structure-based fragment-based ab initio ligand design, based on growing, linking and mutation approaches.
- CombiGlide. Program for ligand-based drug design using ligand-receptor scoring, combinatorial docking algorithms, and core-hopping technology to design focused libraries and identify new scaffolds. Distributed by Schrodinger.
- CoLibri. Assembles huge compound collections from multiple sources and various input formats into a virtual screening library, removes duplicates, assesses the distribution of physico-chemical properties of the compounds and makes selections/filter based on any property-threshold, molecules name-pattern or presence/absence of a particular substructure motif. Generates fragments library. Modifies molecules or fragments for generating, transforming and general handling of virtual screening libraries. Distributed by BioSolveIT.
- ReCore. Replaces a given pre-defined central unit of a molecule (the core), by searching fragments in a 3D database for the best possible replacement, while keeping the rest of the query compound. Additionally, user-defined "pharmacophore" constraints can be employed to restrict solutions. Distributed by BioSolveIT.
- LEGEND. Program for automated structure-based drug design, using an atom-based growing approach. Provided by IMMD.
- Autogrow. Ligand design using fragment-based growing, docking, and evolutionary techniques. AutoGrow uses AutoDock as the selection operator. Provided by the McCammon Group, UCSD.
- CrystalDock. Computer algorithm that aids the computational identification of molecular fragments predicted to bind a receptor pocket of interest. CrystalDock identifies the microenvironments of an active site of interest and then performs a geometric comparison to identify similar microenvironments present in ligand-bound PDB structures. Germane fragments from the crystallographic or NMR ligands are subsequently placed within the novel active site. These positioned fragments can then be linked together to produce ligands that are likely to bind the pocket of interest; alternatively, they can be joined to an inhibitor with a known or suspected binding pose to potentially improve binding affinity. Free and opensource. For Mac OSX, Linux and Windows XP. Developed by the National Biomedical Computation Resource.
- MED-Ligand. Computational fragment-based drug design protocol. Annotated fragments of PDB ligands (MED-Portions) are positioned with MED-SuMo in 3D in a binding site and hybridised with MED-Ligand. Leads are discovered and optimised by hybridisation of MED-Portion chemical moities. Distributed by MEDIT.
- MedChem Studio. Cheminformatics platform for computational and medicinal chemists supporting lead identification and optimization, in silico ligand based design, and clustering/classifying of compound libraries. It is integrated with MedChem Designer and ADMET Predictor. Distributed by Simulation Plus, Inc.
- RACHEL. (Real-time Automated Combinatorial Heuristic Enhancement of Lead compounds). Structure-based all-purpose ligand refinement software package designed to combinatorially derivatize a lead compound to improve ligand-receptor binding. Developed by Drug Design Methodologies and distributed by Tripos.
- MCSS. CHARMm-based method for docking and minimizing small ligand fragments within a protein binding site. Distributed by Accelrys.
- DLD. Automated method for the creation of novel ligands, linking up small functional groups that have been placed in energetically favorable positions in the binding site of a target molecule (See MCSS).
- ACD/Structure Design Suite. Helps chemists optimize the physicochemical properties of their compounds. The software suggests alternative substituents (at a site/sites on the molecule) to drive the property of choice in the desired direction. Helps adjust aqueous solubility, lipophilicity (logP or logD), or change the ionization profile (pKa) of molecules. Distributed by ACD/Labs.
- HSITE. Program for automated structure-based drug design, using fitting and clipping of planar skeleton.
- PRO_LIGAND. Program for automated structure-based drug design, using growing and linking approaches.
- BUILDER. Program for structure-based ab initio ligand design. Finds molecule templates that bind tightly to 'hot spots' in the target receptor, and then generate bridges to join these templates.
- CONCERTS. Program for structure-based ab initio ligand design. Fragments are move independently about a target active site during a molecular dynamics simulation and are linked together when the geometry between proximal fragments is appropriate.
- ADAPT. Program for structure-based ab initio ligand design based pn the DOCK docking software.
- LCT. Program for structure-based ligand design using a linking approach.
- Biogenerator. Program for structure-based design of macrolides using a biomimetic synthesis of substitutide macrolides approach.
- ilib diverse. Program for creating virtual libraries of drug-like organic molecules suitable for rational lead structure discovery. Ligands are designed by combining user-defined fragments according to state-of-the-art chemical knowledge. Generated compounds can be filtered according to a variety of physico-chemical filters. Developed by inte:ligand.
- EMIL. (Example Mediated Innovation for Lead evolution). Suggests chemical modifications to hits to turn them into bona fide leads. EMIL searches through its Knowledge Base looking for similar chemistry and how it was optimized for potency and bioavailability (Iientification of bioisosteric/bioanalogous structures, indication of empirical information of the modification, such as change in physicochemical, in vitro and in vivo effects, etc...). Distributed by CompuDrug.
- Legio. Indigo-based GUI application that exposes the combinatorial chemistry capabilities of Indigo. Free and open source. Distributed by GGA software.
Databases
- SwissBioisostere. Freely available database containing information on millions of molecular replacements and their performance in biochemical assays. It is meant to provide researchers in drug discovery projects with ideas for bioisosteric modifications of their current lead molecule, and to give access to the details on particular molecular replacements. Users can provide a molecular fragment and get possible replacements, along with the biological assays in which they were observed. Users can also provide a given molecular replacement and get the corresponding information. The data were created through detection of matched molecular pairs and mining bioactivity data in the ChEMBL database. Developed and maintained by Merck Serono and the Swiss Institute of BioInformatics.
- VAMMPIRE. (Virtually Aligned Matched Molecular Pairs Including Receptor Environment) matched molecular pairs database for structure-based drug design and optimisation. By building MMPs between PDBbind and ChEMBLdb ligands VAMMPIRE extrapolates the two-dimensional ChEMBLdb ligands to the assumed, three-dimensional binding mode and introduce the received binding information into the database. Provided by the Institute of Pharmaceutical Chemistry / Goethe University Frankfurt, Germany.
- sc-PDB-Frag. Database of protein-bound fragments to help selecting truely bioisosteric scaffolds. The database allows to (i) search fragment among PDB ligands or sketch it; (ii) define similarity rules to retrieve potential bioisosteres; (iii) score bioisosteres according to interaction pattern similarity; (iv) align bioisosteres to the reference scaffold; (v) Visualize the proposed alignment.
- Glide Fragment Library. Set of 441 unique small fragments (1-7 ionization/tautomer variants; 6-37 atoms; MW range 32-226) derived from molecules in the medicinal chemistry literature. The set includes a total of 667 fragments with accessible low energy ionization and tautomeric states and metal and state penalties for each compound from Epik. These can be used for fragment docking, core hopping, lead optimization, de novo design, etc. Provided by Schrödinger.
- FragmentStore. Fragment Store is a database, primarily designed for pharmacists, biochemists, and medical scientists but also researchers working in cognate disciplines like the fragment based drug design. It provides access to information about fragments of compounds with their properties (e.g. charge, hydrophobicity, binding site preferences). It allow the user to do statistical analysis of the fragments properties and binding site preferences. Moreover, the database supports to build an adequate fragment library for fragment based drug design. Provided by the Structural Bioinformatics Group of Charité Berlin.
Web services
- e-LEA3D. Invents ideas of ligand (scaffold-hopping) by the de novo drug design program LEA3D.
- iScreen. Web service for docking and screening the small molecular database on traditional Chinese medicine (TCM) on user's protein. iScreen is also implemented with the de novo evolution function for the selected TCM compounds using the LEA3D genetic algorithm
- 3DLigandSite. Automated method for the prediction of ligand binding sites. Provided by the Imperial London College.
- PASS. Program for tentative identification of drug interaction pockets from protein structure.
- DEPTH. Web server to compute depth and predict small-molecule binding cavities in proteins
- VAMMPIRE-LORD. LORD (Lead Optimization by Rational Design) is a prediction tool based on the VAMMPIRE database (of matched molecular pairs) and using a atom-pair descriptor to represent the substitution environment. It operates on the principle that molecular transformations cause similar effects in similar substitution environments and is therefore able to extrapolate the knowledge of a given substitution effect to any similar system. LORD was implemented as an easy-to-use web server that guides the user step-by-step through the optimization process of a defined lead compound.
ADME Toxicity
Software
- MedChem Studio. Cheminformatics platform for computational and medicinal chemists supporting lead identification and optimization, in silico ligand based design, and clustering/classifying of compound libraries. It is integrated with MedChem Designer and ADMET Predictor. Distributed by Simulation Plus, Inc.